
Newsroom
PTMs Play Critical Role in Regulation of Broad Range of Cellular Processes
Protein post-translational modifications (PTMs) have been increasingly recognized as playing important roles in various biological pathways via regulation of protein function (e.g. phosphorylation and acetylation). Cellular signaling processes heavily rely on reversible PTMs in their capacity to rapidly reprogram individual protein functions. The research group led by Prof. GE Feng at Institute of Hydrobiology of Chinese Academy of Sciences (IHB) performed a systematic investigation on the prokaryotic cell. Their research aimed to characterize the diverse biological functions of PTMs.
Researchers from GE’s lab carried out a global phosphoproteomic analysis on the model cyanobacterium Synechococcus sp. PCC 7002 and diatom P. tricornutum. The provided dataset may help reveal the physiological functions underlying Ser/Thr/Tyr phosphorylation and facilitate the elucidation of the entire signaling networks in cyanobacteria and diatom. Researchers also performed the site-specific phosphoproteome profiling of T. thermophile to unravel the wide-ranging regulatory scope of this modification.
Collaborating with Prof. DENG Jiaoyu from Wuhan Institute of Virology of Chinese Academy of Sciences, researchers performed a global acetylome analysis on M. tuberculosis H37Ra through the combined use of protein/peptide prefractionation, antibody enrichment, and LC-MS/MS analysis. A total of 226 acetylation sites in 137 proteins were found to be involved in various biological processes, such as glycolysis/gluconeogenesis, citrate cycle and fatty acid metabolism. Further functional studies showed that lysine acetylation may play an important role in colony morphology, biofilm formation and tolerance to heat stress in M. tuberculosis H37Ra. More importantly is that it is the first time that lysine acetylation was found to block the immunogenicity.
The findings will not only serve as important resource for the functional analysis of lysine acetylation in M. tuberculosis, but also facilitate the elucidation of the entire metabolic networks in this life-threatening pathogen. Relevant results were published in the Journal of Proteome Research and in Molecular & cellular proteomics.
Related Links:
“Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis”
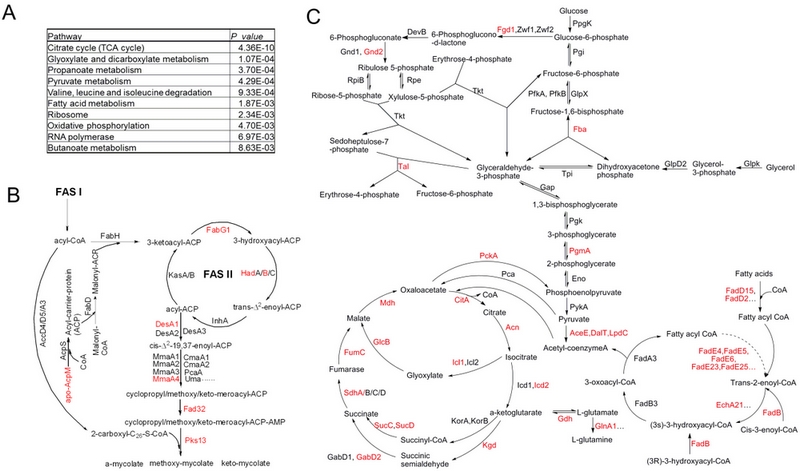 |
| Acetylation of metabolic enzymes inM. tuberculosisH37Ra. (A) Central carbon metabolism network in mycobacteria. (B) FAS-II fatty acid elongation pathway in mycobacteria. Acetylated metabolic enzymes identified by proteomic survey are marked in red. (Image by IHB) |