
Newsroom
Microbial Functions Respond to Damming
As one of the most abundant types of organisms on Earth, bacterioplankton have been found existing in water almost anywhere. They are extremely diverse and highly dynamic, their activities directly influence water conditions, and the biogeochemical cycling.
Bacterioplankton communities also undergo significant shifts in composition or function in response to the environmental variations. The environmental changes from the construction of river dams can greatly affect the aquatic microorganisms living in the reservoir systems. However, most of understanding has focused on community structure, and knowledge regarding microbial function in response to dam is still lacking.
In order to screen the community structure and its function, comprehensive investigation was performed by the research groups led by Dr. YU Yuhe and Dr. HU Zhengyu at Institute of Hydrobiology of Chinese Academy of Sciences (CAS) and Dr. ZHOU Jizhong at Institute for Environmental Genomics of university of Oklahoma to characterize the functional response of bacterioplankton to the damming.
In their study, the metagenomic methods of GeoChip 5.0 and meta-sequencing were applied to address the functional issues of bacterioplankton collected from the Xiangxi River. The results indicated that bacterioplankton communities were both taxonomically and functionally different between backwater and riverine area, which represent communities with and without direct dam effects, respectively.
There were many more nitrogen cycling Betaproteobacteria (e.g., Limnohabitans), and a higher abundance of functional genes and Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) groups involved in nitrogen cycling in the riverine area. This suggests a higher level of bacterial activity involved in generating more nitrogenous nutrients for the growth of phytoplankton. The KO categories involved in carbon and sulfur metabolism, as well as most of the detected functional genes also showed clear backwater and riverine patterns.
Although different methods used in describing the microbial diversity did not necessarily give identical results, the results suggest that the culture-independent approaches used in this study detected similar bacterioplankton patterns along the Xiangxi River. Moreover, all of the diversity revealed by the different methods significantly correlated with the environmental factors. The nearest taxon index (NTI, all values larger than 2) also confirmed that environmental filtering was one of the major processes driving the bacterioplankton community changes in the Xiangxi River.
This work was supported by the China Three Gorges Corporation, the Youth Innovation Promotion Association, CAS and the National Natural Science Foundation of China.
The study was published in Scientific Reports entitled “Impacts of the Three Gorges Dam on microbial structure and potential function”.
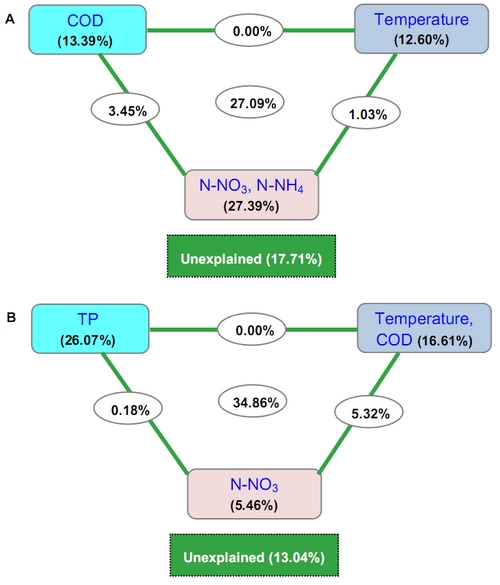 |
| Variance partitioning canonical correspondence analysis (CCA) shows the relative effects of multiple variables on the community composition (A) and potential function (B) of bacterioplankton (Figure by IHB YAN Qingyun and BI Yonghong). |