
Newsroom
Comparative Transcriptome Analyses Provide Insights into Genetic Mechanisms of Adaptation of Tibetan Loaches to High-altitude Environment
The cold climate on the Tibetan Plateau, with hypoxic conditions and strong ultraviolet radiation, creates environments that are extremely hostile to most organisms. As the primary component of the fish fauna on the Tibetan Plateau, Tibetan loaches (Triplophysa fishes) are well adapted to these high-altitude environments. Nevertheless, the genetic mechanisms of the adaptations of these fishes to this high-altitude environment remain poorly understood.
In order to identify the underlying genetic mechanisms for adaptation of fishes to high-altitudes, a research team led by Prof. HE Shunping from Institute of Hydrobiology, Chinese Academy of Sciences (IHB), conducted comparative transcriptome analyses between Triplophysa fishes and fishes living at low altitudes.
They found Triplophysa lineages exhibited the accelerated evolution at the genome scale. Positively selected genes (PSGs) and fast evolving genes (FEGs) were significantly enriched in functional categories related to energy metabolism and to adaptation to hypoxia. The number of Triplophysa lineage-specific nonsynonymous mutations was significantly higher in the FEGs, the PSGs and the overlap of FEGs and PSGs. Selection analysis of the HIF alpha paralogs revealed that HIF-1A and HIF-2B genes experienced positive selection. The positively selected sites were also Triplophysa lineage-specific nonsynonymous mutations and located in functional domains of the protein and predicted functional consequences.
This study is the first report of investigating the genetic mechanisms of adaptation to the conditions on Tibetan Plateau in the Tibetan loaches. The detection of accelerated evolution, the identified candidate genes and the Triplophysa-specific variations related to energy metabolism and hypoxia response provide evidence for adaptations to the high-altitude environment in the Tibetan loaches. The research finding was published online in Genome Biology and Evolution Journal entitled “Evidence for adaptation to the Tibetan Plateau inferred from Tibetan loach transcriptomes” This work was supported by the Pilot projects.
 |
|
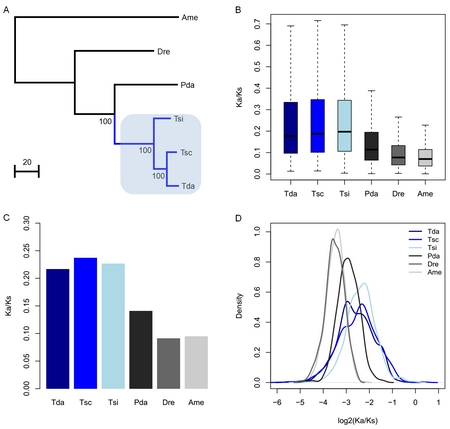
Phylogenetic tree used in this study (A) and the Ka/Ks ratios for the terminal branches obtained from each ortholog (B), concatenated alignments constructed from all orthologs (C), and 1000 concatenated alignments constructed from 10 randomly chosen orthologs (D). The blue line in A represents theTriplophysafishes, which are highlighted in light blue. Tda,T. dalaica; Tsc,T.scleroptera; Tsi,T. siluroides; Pda,Paramisgurnus dabryanus; Dre,Danio rerio; Ame,Astyanaxmexicanus. (Figure by IHB) |