
Newsroom
A Key Regulatory Gene for Bacterial Floc Formation Identified in a Floc-Forming Aquincola Bacterium
Activated sludge (AS) process has been widely applied in the treatment of municipal sewage and industrial wastewater in China and around the globe. However, this classical technology faces many challenges including production of large amounts of excess sludge and the sludge bulking (a phenomenon that sludge flocs float on the surface and could not settle down gravitationally in the clarifier) caused by overgrowth of filamentous bacteria (termed filamentous bacteria bulking) and overproduction of extracellular polysaccharides (viscous bulking). The purification efficiency and effluent quality are undermined as a consequence and more fundamental researches are urgently needed to address such issues.
Previously the research group led by Dr. QIU Dongru from Institute of Hydrobiology (IHB) of Chinese Academy of Sciences (CAS) and their collaborators identified a large gene cluster, including two asparagine synthetase paralogues, required for extracellular polysaccharide biosynthesis and floc formation in the Zoogloea resiniphila, a predominant floc-former isolated from activated sludge in Wuhan and Hong Kong.(An et al., 2016. Water Research, 102: 494-504).
More recently, by using a floc-forming bacterium, Aquincola tertiaricarbonis RN12, as a model, YU Dianzhen, a PhD student and his collaborators have not only identified a similar large gene cluster for biosynthesis of extracellular polysaccharides, but also identified four rpoN paralogues, one of which (rpoN1) is required for floc formation. In addition, this RpoN (sigma54) sigma factor regulates the transcription of genes involved in biofilm formation and swarming motility as previously shown in other bacteria. However, this RpoN paralogue is not required for the biosynthesis of exopolysaccharides, which are released and dissolved into culture broth by the rpoN1 mutant rather than tightly bound to cells as observed during the flocculation of wild-type strain.
These results indicate that floc formation is a tightly regulated complex process and other yet-to-be identified RpoN1-dependent factors are involved in self-flocculation of bacterial cells by exopolysaccharides and/or other biopolymers. The identification and characterization of RpoN-regulated downstream genes required for floc formation are still underway. This study has been published on-line in
This work was supported by Chinese Academy of Sciences and CAS One-Hundred Scholar Award.
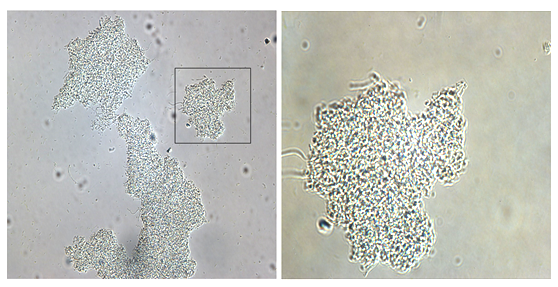
Figure 1 Microscopic observation of bacterial flocs formed by wild-type Zoogloea resiniphila MMB strain. Microphotographs were taken using Nikon Eclipse 80i (Japan) light microscope and a coupled digital camera (Nikon DS-Ri1). Left: 200 times of magnification; Right: 600 times of magnification (An et al., 2016). (Image by IHB)
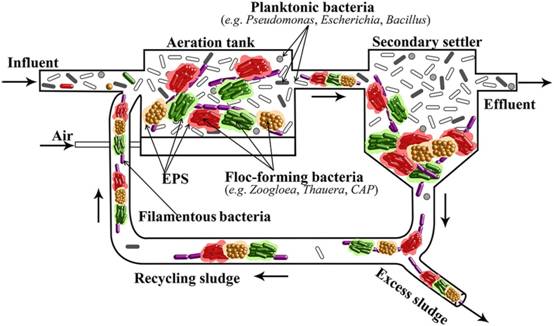
Figure 2 Conceptual model illustrating activated sludge (AS) floc formation of Zoogloea strains and other AS bacteria in activated sludge process (An et al., 2016). Many other crucial AS bacteria including the nitrifying Nitrosomonas strains and polyphosphate-accumulating Candidatus Accumulibacter phosphatis (CAP) harbor a similar extracellular polysaccharide biosynthesis gene cluster and might also be capable of floc formation and thus could be readily enriched for nitrogen and phosphorus removal via the sludge sedimentation and recycling process. (Image by IHB)
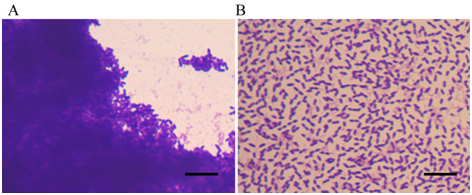
Figure 3 The Aquincola tertiarcarbonis wild-type strain RN12 and a mutant deficient in floc-formation visualized under microscope, stained with crystal violet staining solution (1000 times of magnification, scale bar: 10um). (Image by IHB)