
Newsroom
Scientists Reveal Evolutionary History of Vertebrate Interferon Regulatory Factor
Interferon regulatory factor (IRF) was discovered as transcription factors that regulate the transcription of human interferon (IFN)-β at first. In further studies, more and more IRF members were identified with divers functions. Also, the phylogeny pattern of IRF in invertebrate was found vastly different from that in vertebrate, where advance immune system evolved, detonating the “big bang” vertebrate immune system. Hence to study the phylogeny and functional diversity of IRF not only gains the knowledge about the gene itself, but also may provide new insight into the evolution of immune system.
Lack of key genetic data hinders the phylogeny and function diversity study of IRF. The Research Group of Fish Phylogenetics and Biogeography at Institute of Hydrobiology (IHB) of Chinese Academy of Sciences led by Prof. HE Shunping recently sequenced the genomes and transcriptomes of some ancient fish and lamprey, providing a good insight into the vertebrate ancient genome, hence facilitate the IRF study.
Depending on the similarity of sequence, the researchers identified 148 IRFs in 11 vertebrates and 4 protochordates. Reconstruction of phylogeny tree revealed that the phylogeny pattern of IRF is more complex but stable in vertebrate than in invertebrate. Together with synteny analysis, the results suggest vertebrate IRFs were evolved from 3 predecessors, instead of 4 as suggested in a previous study. The 2nd whole-genome duplication (2WGD) followed by some specific gene duplication expanded the vertebrate IRF members from 3 to 6 and then to 11. Expression analysis results revealed that IRFs evolved from the same predecessors tend to express differently across tissues, indicating a functional diversity.
Even though positivity selection sign has been detected profoundly across vertebrate IRF members, each predecessor tend to have an “offspring” undergoing purifying selection, This "offspring" may hold the ancient function for the predecessor, freeing the others to evolved new functions. Integrating previous studies, the researchers inferred that interferon (IFN), an immune gene new bore in the early evolution of vertebrate, could have greatly contributed the expansion of vertebrate IRFs by introducing new gene interaction and new selection pressure into the existed IRF net work. Furthermore, similar mechanism could also be used extensively in the evolution of vertebrate immune system.
The study was supported by the National Natural Science Foundation of China (Grant No. 31372190). It was published online in Developmental & Comparative Immunology with the title of “Ancient duplications and functional divergence in the interferon regulatory factors of vertebrates provide insights into the evolution of vertebrate immune systems” (link to the paper: https://www.sciencedirect.com/science/article/pii/S0145305X17305633).
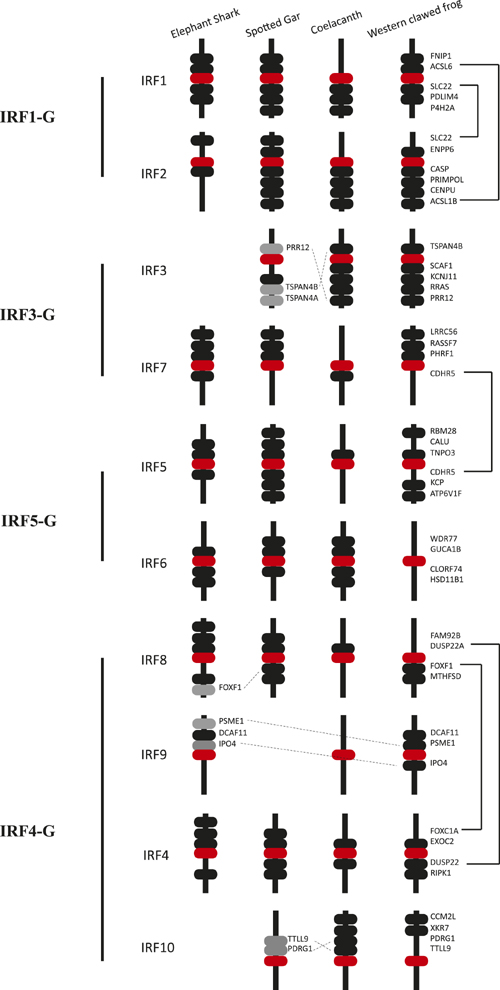
Synteny analysis revealing IRFs and the genes nearby in the genome of representative vertebrate taxa elephant shark, spotted gar, coelacanth and western clawed frog. Blocks in red represent the target IRF gene; blocks in black, the flanked genes echo across species; blocks in grey, the flanked genes that are not in the identical order to that in other species. Vertebrate IRFs were grouped into phylogeny group IRF1-G, IRF3-G, IRF5-G and IRF4-G, each was suggested evolved from a predecessor. However the synteny pattern indicates that group IRF3-G and IRF5-G may evolved from the common predecessor. (Image by IHB)