
Newsroom
How Ten Different Species with Almost Identical Morphology Evolved?
The concept of morphospecies (or cryptic species) is usually applied to a taxonomic species containing multiple morphologically indistinguishable or very similar cryptic biological species. Finding of morphospecies from single-celled protists to the higher animals over the past several decades, make people realized how common morphospecies on our planet. The existence of such a lot of morphospecies challenges our understanding of the taxonomy, biodiversity, ecology, and evolution. However, a little was known for how they are originated and evolved.
Recently, the Research Group of Protozoan Functional Genomics (PI: Prof. MIAO Wei) at Institute of Hydrobiology (IHB) of Chinese Academy of Sciences revealed the hidden genomic evolution behind the ten different species with indistinguishable morphology using a ciliate morphospecies (PLoS Biology, 2019, 17(6): e3000294).
Tetrahymena (Image by IHB)
Focusing on the well-known ciliate morphospecies--Tetrahymena morphospecies, they found that how the microevolution processes operated the genome changes of these morphologically very similar species.
The unique feature of ciliates, separation of micronucleus (germline) and macronucleus (somatic) in a single cell, allow to give a clear picture that what kind of genes (genome innovations) are finally employed by soma, and what kind of elements in germline have important roles to contribute these innovations.
They found that parallel evolutionary trajectories have independently generated a large number of young (species-specific) leucine-rich repeat (LRR) genes with a unique 90-bp exon array in every Tetrahymena species examined. These genes originated at the pericentromeric and subtelomeric regions of chromosomes in the germline genome, and underwent clonal expansions mediated by the retrotransposition of a LINE-like element in concert with unequal crossing-over.
Diverse structures and strong modular organizations for thousands of these LRR genes, somewhat like the CRISPR system in bacteria, seems to have been employed by the somatic genome in evolution, and seems to contribute the environmental responses or ecological adaptation for Tetrahymena species.
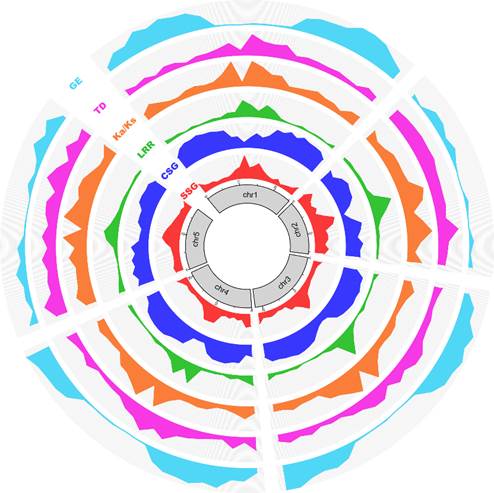
Genomic evolution patterns of Tetrahymena morphospecies (Image by IHB)
This is the first time open a window to the in-depth understanding of the evolution of widespread morphospecies, and has significance for several fields, from genetics, genomics, and evolution to morphospecies biodiversity.
The research was funded by the National Natural Science Foundation of China and Youth Innovation Promotion Association of the Chinese Academy of Sciences. The genomic data of this research has been integrated into the Tetrahymena Comparative Genome Database (TCGD) (Database, 2019, baz029), and provide public web service.