
Newsroom
Study Reveals Two Transcription Factors Interplay in a Cyanobacterium, Improving Adaptation to Changing Environments
Cyanobacteria (also known as blue-green algae) are ancient and the only prokaryotes performing oxygenic photosynthesis. Some filamentous cyanobacteria are capable of heterocyst differentiation to fix and assimilate atmospheric nitrogen when combined nitrogen sources are limiting in the environment. Transcription factors that regulate gene expression play a crucial role in adapting to complex and changing environments.
NtcA, belonging to the cAMP receptor protein (CRP) family, has been extensively studied and well-characterized as a global transcription factor. Despite its pleiotropic and global regulatory capabilities, NtcA is not essential for cyanobacterial survival. DevH, as another CRP family transcription factor, is poorly studied, and its true function remains unknown. How does DevH act? Is it truly essential? And how do these two CRP-family transcription factors interplay in the same organism?
Recently, the research group led by Professor ZHANG Cheng-Cai at the Institute of Hydrobiology (IHB) of the Chinese Academy of Sciences reported in Cell Reportsthe comprehensively characterized function of DevH and its interesting interplay with NtcA.
Professor Zhang's team has long been dedicated to basic research on prokaryotic cell morphogenesis and environmental adaptation, as well as synthetic biology research related to cyanobacterial biotechnology and the control of cyanobacterial bloom. This work is another important achievement of the team after their publication earlier in PNAS, indicating the involvement of DevH in cell morphogenesis.
The present study further demonstrated that the devH gene is essential for cyanobacterial survival under various conditions. Moreover, DevH influences heterocyst differentiation starting from the early stage, rather than only affecting late-stage maturation as previously reported in the literature. By identifying the direct regulon by DevH, this study elucidated the mechanism underlying the essentiality of DevH. As a global transcription factor, it directly regulates various key physiological processes, such as photosynthesis, carbon-nitrogen metabolism, and the cell cycle. Additionally, the conserved DNA-binding motif recognized and bound by DevH was identified to be highly similar to the one for NtcA.
These findings indicate that DevH and NtcA, two CRP-family transcription factors, exhibit substantial functional overlap. However, one is essential for survival, while the other is only essential under specific conditions, suggesting inherent functional specificities. How, then, does the essentiality of DevH emerge compared to NtcA? The team at the end addressed this question by investigating multiple aspects, including their auto-regulatory and cross-regulatory patterns, intracellular protein levels, structural differences, and phylogenetic evolutionary relationships. During evolution, compared with NtcA, DevH in Anabaena has developed distinct auto-regulatory patterns, shows significantly higher protein levels, and undergone functionally meaningful changes in protein structure, such as the acquisition of a positively charged E-loop (which facilitates binding to DNA targets) and the loss of the binding pocket for the small molecule α-ketoglutarate (2-OG). These characteristics enhance DevH’s functions, rendering it an essential transcription factor for cells.
The results of this study suggest that the cooperative interaction between transcription factors of the same family, with both overlapping and specific functions, may represent a key physiological mechanism enabling cyanobacteria to maintain cell growth and survival in dynamically changing environments. Meanwhile, this study also provides a paradigm for exploring functional emergence during evolutionary processes.
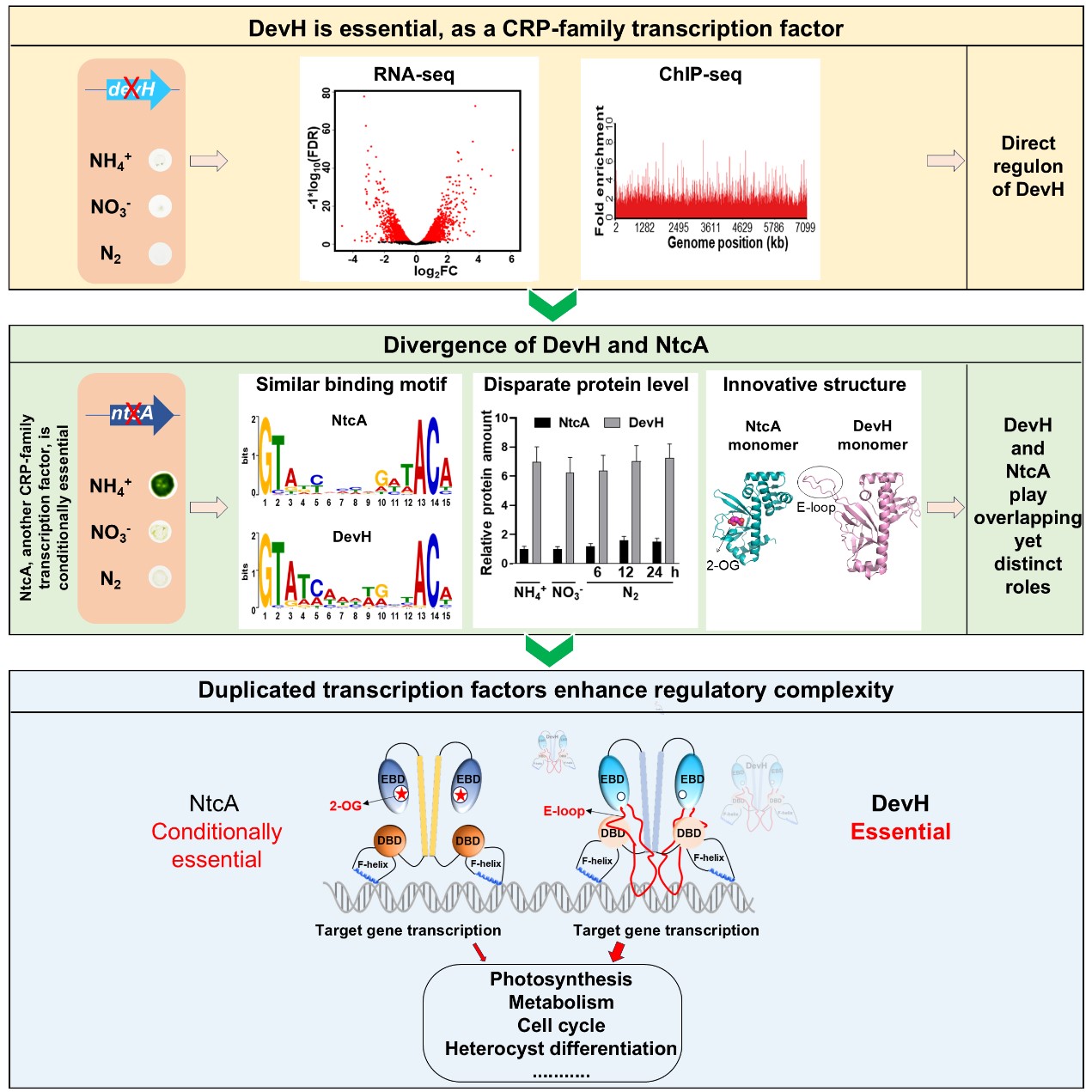
Schematic diagram illustrating the molecular mechanisms underlying the realization and emergence of DevH’s essential functions. (Image by IHB)
(Editor: MA Yun)