Highlights
Protist 10,000 Genomes Project
The Innovation - Protists are a highly diverse group of microscopic eukaryotic organisms that are not fungi, animals, or plants. Protists were some of the microbes first visualized and described by Anton van Leeuwenhoek using the microscope in the seventeenth century. After that, the description and cataloging of these diverse microbial eukaryotes was pursued by microbiologists throughout the following centuries.1 So far, more than 60,000 protist species have been recorded in the NCBI taxonomy system, but many have yet to be identified. Protists have long been considered important models in fundamental biological studies, such as cell biology, genetics, ecology and evolution, toxicology, and applied fields, including biofuels, nutritional supplements and aquaculture feed production, environmental monitoring and pollution treatment, protozoan parasitic disease treatment and prevention, as well as agriculture.
In 1969, Robert Whittaker proposed the popular five-kingdom scheme2 (Figue 1). The protists, which made up the Protista, was regarded as one of the five kingdoms of life (Monera, Protista, Fungi, Plantae, and Animalia). As one of four kingdoms of eukaryotes, only 1,246 (1.9% of total) sequencing projects were performed for 1,151 (3.5% of total) Protista organisms according to the statistics of Genomes Online Database (GOLD). The extreme biased distribution of the current sequencing projects in different kingdoms indicates that there is a huge gap in the understand of the origin, evolution, and diversity of protists.3 Large-scale genome projects have been presented for all the kingdoms of life except the Protista, including the 10,000 Plant Genomes Project (10KP) for plants; the 1000 Fungal Genomes Project (1KFG) for fungi; the 5,000 Arthropod Genomes Project (i5K), Dog 10K Genomes Project (Dog10K), Bird 10,000 Genomes Project (B10K), and Fish 10,000 Genomes Project (FISH10K) for different clades of animals; and the Chinese Academy of Sciences (CAS) Initiative of Microbiome (CMI) and Earth Microbiome Project (EMP) for prokaryotes.
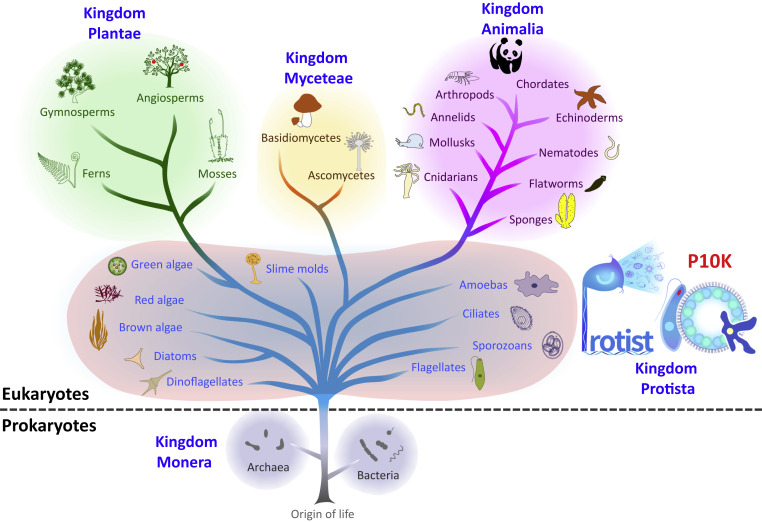
Figure 1. The Protist 10,000 Genomes Project
The logo of P10K (right) is depicted by protists, where a ciliate Vorticella represents the character “P”, combination of a flagellate Euglena and a green algae Volvox represents the number “10”, and an amoeba represents the character “K”. The double helix represents water waves and means the aqueous environments in which the protists live. Small cartoons of protists are ejected from “P”, indicating that many protist species were identified after the Vorticella has been found by Antony van Leeuwenhoek in 1676.
Considering the extremely unbalanced distribution of sequenced eukaryotic genomes and the under-representation of Protista genomes, scientists in six institutions in China, including the Institute of Hydrobiology (IHB), CAS, Tibet University (UTIBET), Henan Agricultural University (HAU), Lanzhou Veterinary Research Institute (LVRI) – Chinese Academy of Agricultural Sciences (CAAS), the Beijing Institute of Genomics (BIG) – CAS, and Huazhong University of Science and Technology (HUST), jointly launched the Protist 10,000 Genomes Project (P10K, Figure 1) on Dec 30, 2019, in Wuhan, China, and thus the P10K consortium was established. The IHB does excellent work in phycology and protozoology, and runs the National Aquatic Biological Resource Center (NABRC) in China, which contains more than 3,000 strains of protist. The UTIBET conducts long-term research of protozoa with special interests in Qinghai-Tibet Plateau. The HAU and LVRI mainly focuses on the parasitic protists, especially for livestock and poultry, and run part of the National Parasitic Resource Center (NPRC) in China. The BIG is an excellent institution in genomics and runs the National Genomics Data Center (NGDC) of China, which supports data analysis and management. The HUST has excellent teams in metagenomics and supports protist genomic data mining from metagenomics data.
The primary goals of P10K are to generate the whole-genome sequences (WGSs) of 10,000 protists and create new reference genomes for the representative species of different lineages of protists. The rapid development of genome sequencing technologies (in particular, single-cell sequencing of protists that are not amenable to laboratory culture) combined with bioinformatic technologies on algorithms (in particular, data mining of metagenomes) and databases has made it feasible to decipher the genome sequences of protists, reconstruct their phylogenetic trees, and accordingly address fundamental scientific questions of broad interest, such as the origin of eukaryotes and multicellularity, the diversification of eukaryotes, the adaptive evolution of protists toward the extreme conditions, microbe interaction, and genetic resources mining.
P10K will be the first international genome sequencing initiative focusing on the kingdom of Protista. P10K will run as an open initiative. We invite teams worldwide that could contribute the samples, sequencing, and bioinformatics analysis to join P10K. At the time of this writing, a team from Jilin Agriculture University focusing on slime molds, and a team from Southwest University focusing on microsporidia, have joined this project.
References
1. Haeckel, E. (1899). Kunstformen der Natur. In Die einhundert Farbtafeln im Faksimile mit beschreibendem Text, allgemeiner Erlauterung und systematischer € übersicht, 2. (Leipzig and Vienna: Bermann-Fischer), pp. 1899–1904.
2. Whittaker, R.H. (1969). New concepts of kingdoms of organisms. Science 163, 150–160.
3. del Campo, J., Sieracki, M.E., Molestina, R., et al. (2014). The others: our biased perspective of eukaryotic genomes. Trends Ecol. Evol. 29, 252–259.