Highlights
CRP Paralogues Leverage Dissimilatory Nitrate Reduction to Ammonium and Denitrification Pathways for Maximizing Energy Generation and Bacterial Growth under Different Ambient Carbon/Nitrogen Ratios
Based on ambient carbon/nitrogen (C/N) ratios, researchers led by Prof. QIU Dongru from Institute of Hydrobiology (IHB) of Chinese Academy of Sciences recently revealed that cyclic AMP receptor protein (CRP)1, which is widely encoded in bacteria, is required for Dissimilatory Nitrate Reduction to Ammonium (DNRA) in Shewanella loihica PV-4 strain, while the CRP2 paralogue is required for transcription of the nitrite reductase gene nirK for denitrification.
The paper has been published online in Applied and Environmental Microbiology.
Large-scale industrial nitrogen fixation and chemical fertilizer application have not only dramatically increased crop yields to feed the growing population, but enhanced the reactive nitrogen input into soils, surface and ground waters, and accelerated the eutrophication of freshwaters and coastal marine environments. The increasing emission of potent greenhouse gases including nitrous oxide may have strikingly aggravated global warming and climate change. Some microbes utilize different dissimilatory nitrate reduction (DNR) pathways, including DNRA and denitrification pathways, for anaerobic respiration in response to ambient carbon/nitrogen ratio changes. However, little is known about the molecular mechanism underlying the choice of two competing DNR pathways.
By performing a series of molecular genetics and analytical chemistry experiments, IHB Researchers found that deletion of crp1 could accelerate the reduction of nitrite to NO under both low and high C/N ratios. CRP1 is not required for denitrification and actually suppresses production of NO and N2O gases. Deletion of either of the NO-forming nitrite reductase genes nirK or crp2 blocked production of NO gas.
Furthermore, real-time PCR and electrophoretic mobility shift assays (EMSAs) demonstrated that the transcription levels of DNRA-relevant genes such as nap-β (napDABGH), nrfA, and cymA were upregulated by CRP1, while nirK transcription was dependent on CRP2.
It is clear that both CRP paralogues are involved in the bacterial choice of two competing dissimilatory nitrate reduction pathways, DNRA and denitrification. Sufficient carbon source leads to the predominance of DNRA for more energy per mol nitrate as electron acceptor and ammonia available for amino acid synthesis, while denitrification is favored for more energy per mol lactate as electron donor and saving carbon sources for bacterial growth under low C/N ratios.
Nevertheless, carbon source/electron donor deficiency may result in an incomplete denitrification process, raising the concern of high levels of N2O emission from nitrate-rich and carbon source-poor waters and soils.
The findings not only shed light on important insights into the understanding of microbe-driven biogeochemistry cycles of nitrogen, but provide important implications for efficient control of emission of N2O to the atmosphere from both the hydrosphere and the soil sphere as well.
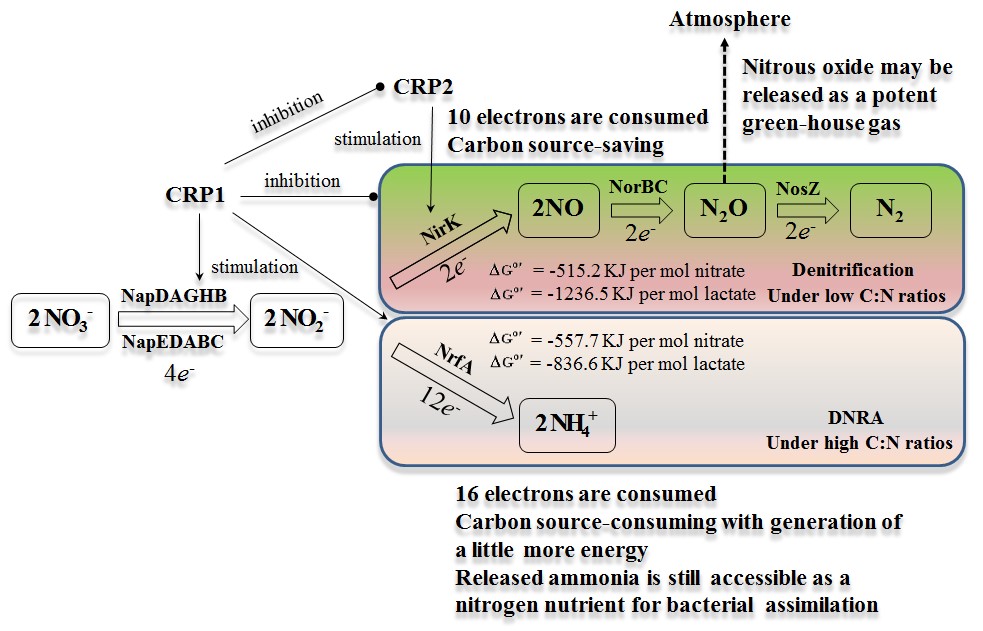
Schematic diagram of the regulation of CRP1 on the dissimilatory nitrate reduction to ammonia and denitrification (Credit: IHB)