Highlights
Regulation of 3D Chromatin Organization is Involved in Feeding Preference Adaptation
One of the primary mechanisms that result in the emergence of new species is divergence in feeding preference, yet little is known about the genetic mechanism that underlie feeding preference adaptation, particularly when it comes to the regulation of the three-dimensional (3D) chromatin organization.
Two sister species, Gymnocypris eckloni scoliostomus (GS) and G. eckloni eckloni (GE), are distributed in the Lake Sunmcuo on the Qinghai-Tibet Plateau in China. GS and GE evolved diverse feeding preferences and demonstrated significant adaptive divergence in the morphology and function of feeding organs despite the fact that they only split about 57,000 years ago.
Recently, a research group led by Prof. HE Shunping from the Institute of Hydrobiology (IHB) of the Chinese Academy of Sciences demonstrated how the regulation of 3D chromatin organization involved in feeding preference adaptation. This study was published in Journal of Genetics and Genomics.
In this study, the researchers combined Hi-C and RNA-seq to explore the relationship between 3D genome conformation and gene expression. They found that A/B compartments are mostly conserved with only 17.5% of compartment A/B switched between GS and GE. Overall, genes that switched from GS-A to GE-B tend to show significantly decreased expression levels, but genes that switched from GS-B to GE-A showed no significant difference in gene expression. These transition regions from GS-A to GE-B are mainly enriched in lipid metabolism-related pathways, highlighting the possibility of active expression of these genes in GS's liver tissue.
What’s more, the researchers found only a small percentage of topologically associating domain (TAD) and DNA loops were completely conserved, and the genes within species-specific TADs showed significant expression differences when compared to genes in conserved TADs. These species-specific TADs are often accompanied by alterable local DNA loop interaction. These results suggested that TAD and DNA loop play an important role in regulating gene expression.
The researcher further identified nine candidate genes of lipid metabolism. Interestingly, eight of the nine candidate genes underwent TAD structural rearrangements or DNA loop rearrangements. They also found that the TAD and DNA loop rearrangement likely played an instructive role in regulating gene expression by demarcating TAD structures and modifying the specificity of enhancer/promoter interactions. The TAD boundary may determine the regions where enhancer/promoter interactions are facilitated or insulated.
In summary, this study provides a new 3D epigenomic perspective for understanding the evolution of different feeding preferences adaptation.
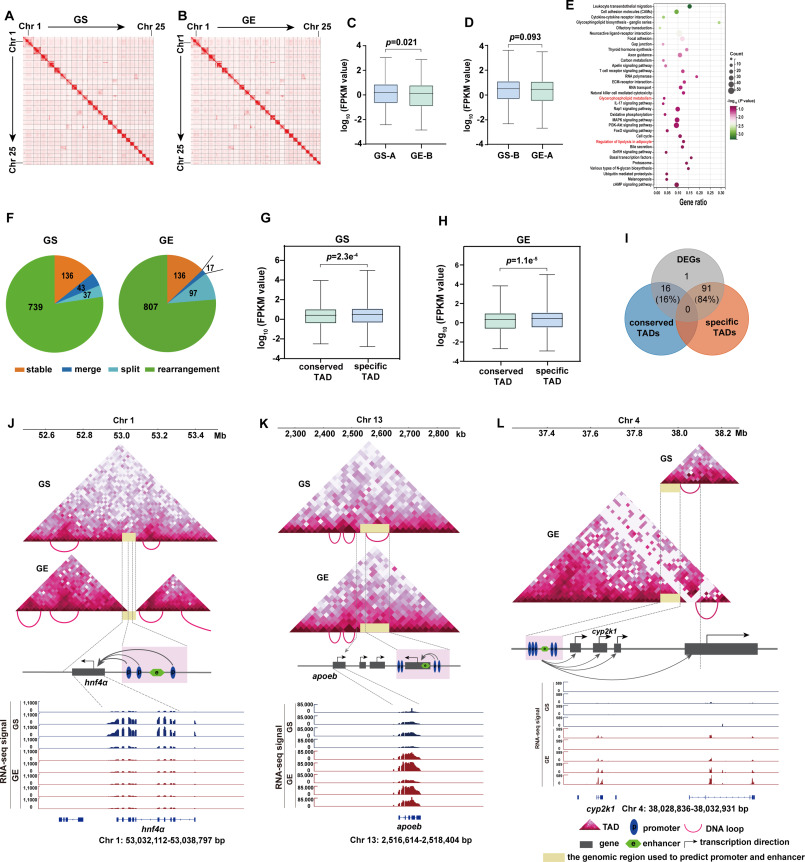
Genome-wide chromatin structure changes and gene expression between Gymnocypris eckloni scoliostomus (GS) and G. eckloni eckloni (GE). (Image by IHB)
(Editor: MA Yun)