Highlights
Genome of Chinese Longsnout Catfish Provides Novel Insights into Carnivore Feeding Preferences and Corresponding Metabolic Strategies
Fish exhibits diverse feeding habits as adaptations to their complex living environments. Over millions of years of evolution, fish have developed distinct anatomical features related to feeding habits and food use. These habits include a carnivorous diet, a herbivorous diet, and an omnivorous diet. Recent studies have revealed the roles of olfactory and gustatory organs in fish feeding preferences, which contribute to their dietary adaptation. However, the genetic basis of feeding preference and the corresponding metabolic strategies that differentiate feeding habits remain elusive.
The Chinese longsnout catfish (Leiocassis longirostris Günther) serves as an excellent carnivorous fish model, characterized by low efficiency in food carbohydrate utilization and a high demand for food protein content (≥ 45%). Grass carp, a typical herbivorous fish, shifts from a carnivore of feeding on plankton to an herbivore during the larval and juvenile stages. The dietary shift from carnivore to herbivore in grass carp provides an excellent model for understanding the genetic basis of dietary preferences between fish with different feeding habits by comparing its genome with that of other carnivorous fish.
A research group led by Prof. XIE Shouqi from the Institute of Hydrobiology (IHB) of the Chinese Academy of Sciences successfully completed the whole genome sequencing of Chinese longsnout catfish, and revealed a conserved adaptations between fish feeding preferences and metabolic strategies. This study was published in Genome Research.
In this study, the researchers compared the whole genome of Chinese longsnout catfish with that of grass carp, and identified 250 genes through joint analysis of positive selection and rapid evolution, including taste receptor taste receptor type 1 member 3 (tas1r3) and trypsin. Compared to the grass carp, the Chinese longsnout catfish exhibited significant feeding preferences (tas1r3-calhm-cck signaling) and corresponding protein metabolism characteristics (trypsin-lap3-cars2 signaling). In contrast, the grass carp mainly relied on the carbohydrate metabolism pathway (slc2a9-ppp1r3g-chrebp signaling).
Further studies revealed that the tas1r3 gene plays a key role in the evolution of feeding habits in carnivorous fish. It was confirmed that tas1r3 is necessary for carnivore preference in tas1r3-deficient zebrafish and in a diet-shifted grass carp model. Additionally, marked alterations in trypsin activity and metabolic profiles are accompanied by a transition of feeding preference in tas1r3-deficient zebrafish and diet-shifted grass carp.
The study not only reveals a conserved adaptation between feeding preference and corresponding metabolic strategies in fish, but also provide novel insights into the evolution of feeding habits and metabolism in fish.
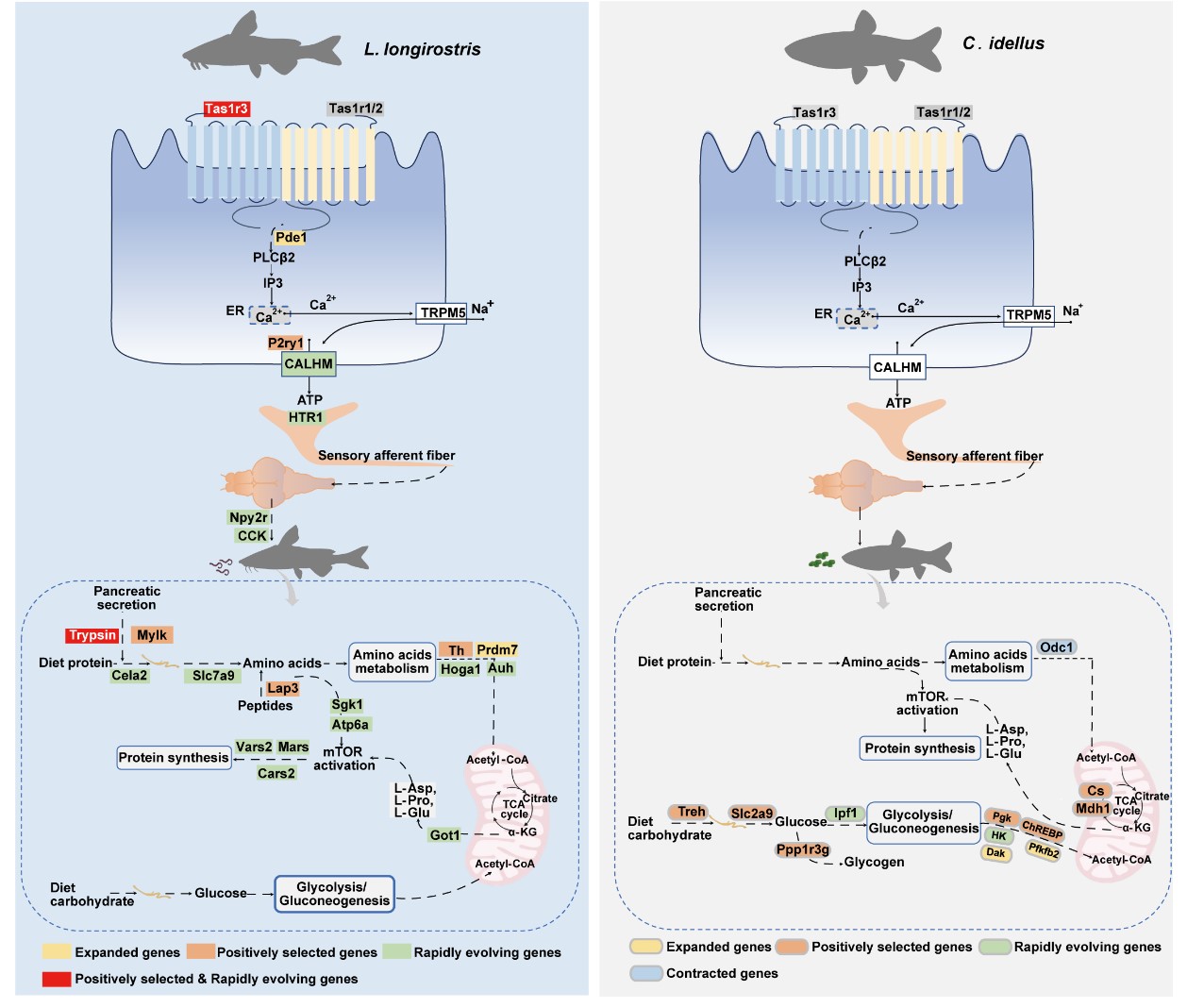
Comparative Genomic Analysis of Chinese Longsnout Catfishand Grass Carp (Image by IHB)
(Editor: MA Yun)