
Newsroom
A Series of Advances in the Sexual Reproduction of Protozoa
Sexual reproduction plays an essential role in the propagation and evolution of eukaryotes, establishing a remarkable spectrum of diversity across various species. While investigations into the regulation of sexual reproduction have been undertaken in eukaryotes spanning animals, plants, and fungi, comprehensive and thorough analyses of the molecular mechanisms governing sexual reproduction remain confined to a few organisms.
Tetrahymena thermophila, a model protozoan with seven mating types, represents one of the early branched unicellular eukaryotes. Its unique characteristics and evolutionary positioning present a good opportunity to systematically delve into the regulatory intricacies of sexual reproduction within early differentiated eukaryotic taxa. Furthermore, it offers a platform to explore the origins and evolutionary underpinnings of sexual reproduction in eukaryotes.
Recently, a research group led by Prof. Wei Miao from the Institute of Hydrobiology (IHB) of the Chinese Academy of Sciences got a series of advances in the study of sexual reproduction of the protozoan T. thermophila, including sexual-reproduction initiation, mating-type recognition and meiotic homologous recombination. These studies were published in PNAS, eLife and Journal of Genetics and Genomics, respectively.
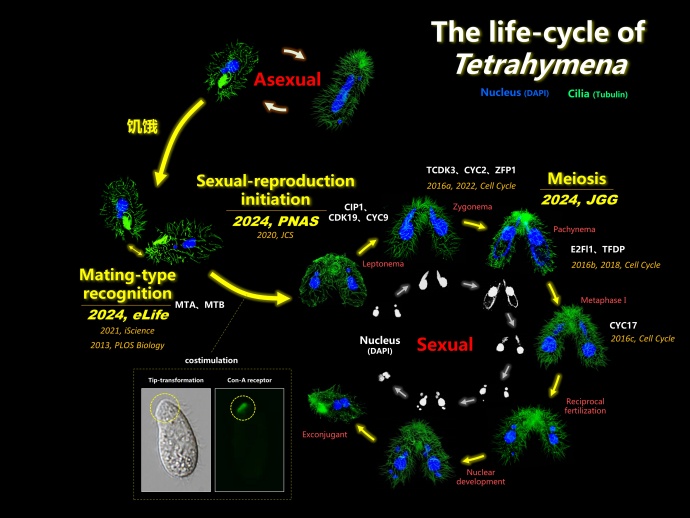
Figure 1. The life-cycle of Tetrahymena
1) How to do sex before the occurrence of sexes?
One of the hallmarks of sexual reproduction in eukaryotes is meiosis. Because the meiosis-related genes are ubiquitous and conserved in eukaryotes, it is suggested that meiosis originated early and already existed in the common ancestor of eukaryotes. However, the sex determining genes adopted by different organisms are quite different, indicating that sex arose relatively late in evolution. However, it is not clear how eukaryotes perform sexual reproduction in the intermediate state from the origin of meiosis to the emerge of mating type. The related hypothesis is that during this period, eukaryotes are unisexual, that is, the pairing of homologous cells (selfing). However, the evidence is lacking to support this hypothesis.
Prof. Wei Miao and his team recently investigated the initiation of sexual reproduction in Tetrahymena and found that deletion of a single gene, CIP1, allows the species to skip mating type recognition and directly initiate sexual reproduction. Deletion of CIP1 resulted in loss of self-avoidance function in mating-type recognition. CIP1 blocks the initiation of sexual reproduction by inhibiting the CDK-cyclin complex (CDK19-CYC9), being responsible for coupling the initiation of sexual reproduction with mating-type recognition. This result supports that the ancestors of eukaryotes are unisexual for sexual reproduction.
This work is published in PNAS entitled “Cip1, a CDK regulator, determines heterothallic mating or homothallic selfing in a protist”. The co-first authors are Dr. Yang Ma and Dr. Guanxiong Yan. The corresponding author is Prof. Wei Miao.
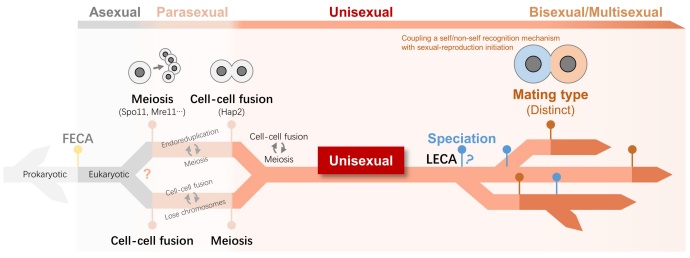
Figure 2. A hypothesized pathway of origin of sexual reproduction
2) How do single-celled organisms recognize same or different sexes?
While most common organisms exhibit only two sexes (female/male,+/-, a/α, etc.), species with multiple sexes are also exist in nature, such as some ciliates and basidiomycetes. The model ciliate Tetrahymena thermophila has seven mating types (I-VII). In earlier research, Prof. Wei Miao and his team, in collaboration with Prof. Eduardo Orias from the University of California, Santa Barbara, discovered that the molecular basis of these seven mating types is MTA and MTB proteins. However, the mechanism underlying the recognition among these seven mating types remains unresolved.
Recently, through collaboration with Prof. Ping Yin's team from Huazhong Agricultural University, it was found that during Tetrahymena mating type recognition, MTA and MTB do not simply engage in ligand-receptor interactions, but rather form a large complex with a series of proteins to exert their function. This complex is located on the cell surface rather than the ciliary surface and is arranged around the cilia in a row. On one hand, this complex can recognize same mating type, thereby inhibiting pairing, and on the other hand, it can recognize different mating types, thereby activating pairing, demonstrating its ability to simultaneously recognize self and non-self.
This work is published in eLife entitled “A seven-sex species recognizes self and non-self mating-type via a novel protein complex”. The first author is Dr. Guanxiong Yan. The corresponding authors are Prof. Wei Miao and Prof. Ping Yin.
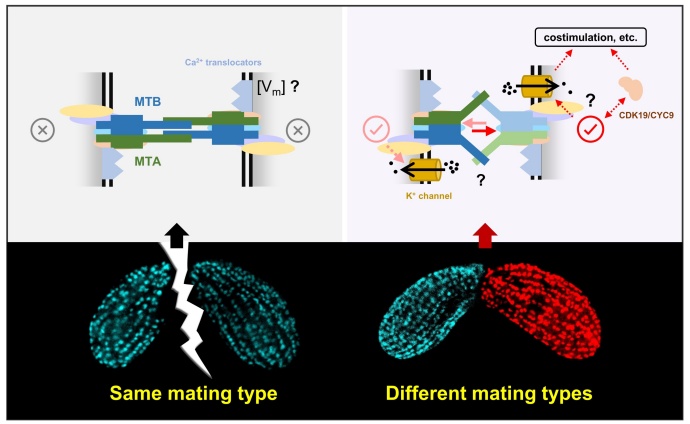
Figure 3. Mating-type recognition model of Tetrahymena
3) What is the meiotic homologous recombination landscape in organisms without synaptonemal complex?
Meiotic homologous recombination is an important process of sexual reproduction, which not only ensures the normal allocation of the parental chromosomes to the gametes, but also affects genome diversity and adaptive evolution of organisms. However, little is known about the patterns of meiotic recombination in ancient evolutionary clades such as the unicellular protozoa ciliates.
Using a high density of single nucleotide polymorphic sites (3.8 SNPs/kb) as molecular markers, Prof. Wei Miao and his team detected 1,021 crossover events in the 38 hybrid progeny of the model ciliate T. thermophila, corresponding to a genomic recombination rate of 9.9 cM/Mb. The recombination rate was much higher than those of human, mouse, Arabidopsis and other higher animals and plants. However, gene conversion events were rare, with only 1.03 per meiosis, and there was no G/C-biased conversion. No obvious sign of crossover interference was found, which might be related to the lack of synaptonemal complex during the meiosis of T. thermophila. In addition, recombination hotspots often occur in centromere or subtelomeric regions and are closely associated with genes that respond to environmental changes. This study sheds light on the pattern of meiotic homologous recombination in synaptonemal complex-deficient species, providing new insights into the regulation of crossover and the evolution of recombination machinery in different taxa.
This work is published in JGG entitled “The genome-wide meiotic recombination landscape in ciliates and its implications for crossover regulation and genome evolution”. The first author is Dr. Lu Fu. The corresponding authors are Prof. Wei Miao and Dr. Guangying Wang.

Figure 4. The cover of Journal of Genetics and Genomics (Volume 51 Issue 3, 2024)
(Editor: WANG Hongxia)